염색체연구소
-
연구소장 소개

안녕하십니까? 염색체연구소를 방문해 주셔서 감사합니다.
염색체연구소장 김현희- •성명 : 김현희
- •이메일 : kimhh@syu.ac.kr
- •소속 : 삼육대학교 생명과학과 교수
연구소 홈페이지
논문
- •Remnyl Joyce Pellerin, Nomar Espinosa Waminal, Hadassah Roa Belandres, and Hyun Hee Kim, Karyotypes of Three Exotic Cucurbit Species Based on Triple-Color FISH Analysis, Korean J. Hortic. Sci. Technol. 36(3):417-425, 2018
- •Nam-Hoon Kim, Murukarthick Jayakodi, Sang-Choon Lee, Beom-Soon Choi, Woojong Jang, Junki Lee, Hyun Hee Kim, Nomar E. Waminal, Meiyappan Lakshmanan, Binh van Nguyen, Yun Sun Lee, Hyun-Seung Park, Hyun Jo Koo, Jee Young Park, Sampath Perumal, Ho Jun Joh, Hana Lee, Jinkyung Kim, In Seo Kim, Kyunghee Kim, Lokanand Koduru, Kyo Bin Kang, Sang Hyun Sung, Yeisoo Yu, Daniel S. Park, Doil Choi, Eunyoung Seo, Seungill Kim, Young-Chang Kim, Dong Yun Hyun, Youn-Il Park, Changsoo Kim, Tae-Ho Lee, Hyun Uk Kim, Moon Soo Soh, Yi Lee, Jun Gyo In, Heui-Soo Kim, Yong-Min Kim, Deok-Chun Yang, Rod A. Wing, Dong-Yup Lee, Andrew H. Paterson,and Tae-Jin Yang, Genome and evolution of the shade-requiring medicinal herb Panax ginseng, Plant Biotechnology Journal 16:1904~1917, 2018, doi: 10.1111/pbi.12926
- •Nomar Espinosa Waminal, Remnyl Joyce Pellerin, Nam-Soo Kim, Murukarthick Jayakodi, Jee Young Park, Tae-Jin Yang, Hyun Hee Kim, Rapid and Efficient FISH using Pre-Labeled Oligomer Probes, Scientific Reports 8:8224, 2018, DOI:10.1038/s41598-018-26667-z
- •Hui Chao Zhou, Eung Jun Park, Hyun Hee Kim, Analysis of Chromosome Composition of Gastrodia elata Blume by Fluorescent in situ Hybridization using rDNA and Telomeric Repeat Probes, Korean J. Medicinal Crop Sci. 26(2) : 113~118, 2018
- •Sung Min Youn, Hyun Hee Kim, Chromosome Karyotyping of Senna covesii and S. floribunda based on Triple-color FISH Mapping of rDNAs and Telomeric Repeats, Plant Breed. Biotech. 6(1):51~56, 2018
- •Nomar Espinosa Waminal, Remnyl Joyce Pellerin, Woojong Jang, Hyun Hee Kim, Tae-Jin Yang, Characterization of Chromosome‑pecific Microsatellite Repeats and Telomere Repeats Based on Low Coverage Whole Genome Sequence Reads in Panax ginseng, Plant Breed. Biotech. 6(1):74~81, 2018
- •Remnyl Joyce Pellerin, Nomar Espinosa Waminal, HyunHee Kim, Triple-color FISH Karyotype Analysis of Four Korean Wild Cucurbitaceae Species, Korean J. Hortic. Sci. Technol. 36(1):98~107, 2018
- •Sampath Perumal, Nomar Espinosa Waminal, Jonghoon Lee, Junki Lee, Beom-Soon Choi, Hyun Hee Kim, Marie-Angèle Grandbastien, Tae-Jin Yang, Elucidating the major hidden genomic components of the A, C, and AC genomes and their influence on Brassica evolution, Scientific Reports 7, 2017
- •Remnyl Joyce Pellerin, Nomar Espinosa Waminal, Ji Young Kim, Yurry Um, Hyun Hee Kim, Fluorescence in situ Hybridization Karyotype Analysis of Seven Platycodon grandiflorum (Jacq.) A. DC. Cultivars, Horticultural Science and Technology 35(6):784~792, 2017
- •Soo-Seong Lee, Byoung Ho Hwang, Tae Yoon Kim, Jeongmin Yang, Na Rae Han, Jongkee Kim, Hyun Hee Kim, Hadassah Roa Belandres, Developing Stable Cultivar through Microspore Mutagenesis in ×Brassicoraphanus koranhort, Inter-Generic Allopolyploid between Brassica rapa and Raphanus sativus, American Journal of Plant Sciences 8: 1345-1356, 2017
- •Nomar Espinosa Waminal, Hong-Il Choi, Nam-Hoon Kim, Woojong Jang, Junki Lee, Jee Young Park, Hyun Hee Kim, Tae-Jin Yang, A refined Panax ginseng karyotype based on an ultra-high copy 167-bp tandem repeat and ribosomal DNAs, J Ginseng Res 41: 469-476, 2017
- •Nomar Espinosa Waminal, Sampath Perumal, Jonghoon Lee, Hyun Hee Kim, Tae-Jin Yang, Repeat Evolution in Brassica rapa (AA), B. oleracea (CC), and B. napus (AACC) Genomes, Plant Breed. Biotech. 4:107-122, 2016
- •Yun Sun Lee, Hye Mi Park, Nam-Hoon Kim, Nomar E. Waminal, Yeon Jeong Kim, Ki-Byung Lim, Jin Hong Baek, Hyun Hee Kim, Tae-Jin Yang, Phylogenetic relationship of 40 species of genus Aloe L. and the origin of an allodiploid species revealed by nucleotide sequence variation in chloroplast intergenic space and cytogenetic in situ hybridization, Genet Resour Crop Evol 63: 235-242, 2016
- •Abigail Rubiato Cuyacota, So Youn Wonb, Sang Kun Parkc, Seong-Han Sohn, Jung ho Lee, Jung Sun Kim, Hyun Hee Kim, Ki-Byung Lim, Yoon-Jung Hwang, The chromosomal distribution of repetitive DNA sequences in Chrysanthemum boreale revealed a characterization in its genome, Scientia Horticulturae 198:438–444, 2016
- •Nomar Espinosa Waminal and Hyun Hee Kim, FISH karyotype analysis of four wild Cucurbitaceae species using 5S and 45S rDNA probes and the emergence of new polyploids in Trichosanthes kirilowii Maxim, Korean J. Hortic. Sci. Technol. :869-876, 2015
- •Nomar Espinosa Waminal, Sampath Perumal, Ki-Byung Lim, Beom-Seok Park, yun Hee Kim, and Tae-Jin Yang, Genomic survey of the hidden 7 components of the B. rapa genome, In: X. Wang and C. Kole (eds.), The Brassica rapa Genome, Compendium of Plant Genomes, pp 83-96, Springer-Verlag, Berlin Heidelberg, 2015
- •Hadassah Roa Belandres, Nomar Espinosa Waminal, Yoon-Jung Hwang, Beom-Seok Park, Soo-Seong Lee, Jin Hoe Huh, and Hyun Hee Kim, FISH Karyotype and GISH Meiotic Pairing Analyses of a Stable Intergeneric Hybrid xBrassicoraphanus Line BB#5, Kor. J. Hort. Sci. Technol. 33(1):83-92, 2015
- •Shengyi Liu, Yumei Liu, Xinhua Yang, Chaobo Tong, David Edwards, Isobel A.P. Parkin, Meixia Zhao, Jianxin Ma, Jingyin Yu, Shunmou Huang, Xiyin Wang, Junyi Wang, Kun Lu, Zhiyuan Fang, Ian Bancroft, Tae-Jin Yang, Qiong Hu , Xinfa Wang, Zhen Yue, Haojie Li, Linfeng Yang, Jian Wu, Qing Zhou, Wanxin Wang, Graham J. King, J. Chris Pires, Changxin Lu, Zhangyan Wu, Perumal Sampath, Zhuo Wang, Hui Guo, Shengkai Pan, Limei Yang, Jiumeng Min, Dong Zhang, Dianchuan Jin, Wanshun Li, Harry Belcram, Jinxing Tu, Mei Guan, Cunkou Qi, Dezhi Du, Jiana Li, Liangcai Jiang, Jacqueline Batley, Andrew G. Sharpe, Beom-Seok Park, Pradeep Ruperao, Feng Cheng, Nomar Espinosa Waminal, Yin Huang, Caihua Dong, Li Wang, Jingping Li, Zhiyong Hu, Mu Zhuang, Yi Huang , Junyan Huang, Jiaqin Shi, Desheng Mei, Jing Liu, Tae-Ho Lee, Jinpeng Wang, Huizhe Jin, Zaiyun Li, Xun Li, Jiefu Zhang, Lu Xiao, Yongming Zhou, Zhongsong Liu, Xuequn Liu, Rui Qin, Xu Tang,Wenbin Liu, Yupeng Wang, Yangyong Zhang, Jonghoon Lee, Hyun Hee Kim, France Denoeud, Xun Xu, Xinming Liang, Wei Hua, Xiaowu Wang, Jun Wang, Boulos Chalhoub & Andrew H. Paterson, The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes, Nature Communications 5(3930), doi:10.1038/ncomms4930, 2014
- •Ji Yeon Kwon, Jin Sung Hong, Min Jea Kim, Sun Hee Choi, Byeong Eun Min, Eun Gyeong Song, Hyun Hee Kim, Ki Hyun Ryu, Simultaneous multiplex PCR detection of seven cucurbit-infecting viruses, Journal of Virological Methods 206:133–139, 2014
- •Hong-Il Choi, Nomar E. Waminal, Hye Mi Park, Nam-Hoon Kim, Beom Soon Choi, Minkyu Park, Doil Choi, Yong Pyo Lim, Soo-Jin Kwon, Beom-Seok Park, Hyun Hee Kim and Tae-Jin Yang, Major repeat components covering one-third of the ginseng (Panax ginseng C.A. Meyer) genome and evidence for allotetraploidy, The Plant Journal 77:906-916, 2014
- •Nomar Espinosa Waminal, Kwang Bok Ryu, Bo Reum Park, Hyun Hee Kim, Phylogeny of Cucurbitaceae species in Korea based on 5S rDNA non-transcribed spacer, Genes Genom 36:57–64, 2014
- •Nomar Espinosa Waminal, Ki Hyun Ryu, Sun-Hee Choi and Hyun Hee Kim, Randomly detected genetically modified(GM) maize(Zea mays L.) near a transport route revealed a fragile 45S rDNA phenotype, Plos One 8(9):1-14, 2013
- •Sang Eun Han, Hyun Hee Kim and Kweon Heo, A cytotaxonomic study of Astragalus koraiensis Y.N. Lee, Korean J. Pl. Taxon. 43(2):139-145, 2013
- •Nomar Espinosa Waminal, Hye Mi Park, Kwang Bok Ryu, Joo Hyung Kim, Tae Jin Yang, Hyun Hee Kim, Karyotype analysis of Panax ginseng C.A. Meyer, 1843(Araliaceae) based on rDNA loci and DAPI band distribution, Comparative Cytogenetics 6(4):425-441, 2012
- •Nomar Espinosa Waminal and Hyun Hee Kim, Dual-color FISH karyotype and rDNA distribution analyses on four Cucurbitaceae, Hort. Environ. Biotechnol. 53(1):60~67, 2012
- •Jae-Han Son, Kyong-Cheul Park, Sung-Il Lee, Eun-Jin Jeon, Hyun-Hee Kim and Nam-Soo Kim, Sequence variation and comparison of the 5S rRNA sequences in Allium species and their chromosomal distribution in four Allium species, J. Plant Biol. 55(1):15-25, 2012
- •Eun Jin Jeon , Kwang Bok Ryu, Hyun Hee Kim, Karyotype Analyses of a rice Cultivar “Nakdong” and its four Genetically Modified Events by Conventional Staining and Fluorescence in situ Hybridization, Kor. J. Breed. Sci. 43(4):173-180, 2011
- •S-H Jo, H-M Park, S-M Kim, HH Kim, C-G Hur and D Choi, Unraveling the sequence dynamics of the formation of genus-specific satellite DNAs in the family Solanaceae, Heredity 106:876-885, 2011
- •Nomar Espinosa Waminal, Nam-Soo Kim and Hyun Hee Kim, Dual-color FISH karyotype analyses using rDNAs in three Cucurbitaceae species, Genes & Genomics 33:521-528, 2011
- •Yoon-Jung Hwang, Hyun Hee Kim, Jong-Bo Kim and Ki-Byung Lim, Karyotype analysis of Lilium tigrinum by FISH, Hort. Environ. Biotechnol. 52(3):292-297, 2011
- •Sung Nam Lee, Sun Hee Choi, Kwang Bok Ryu, Hyun Hee Kim and Ki Hyun Ryu, Molecular and Cytogenetic assessment of transgenic hot peppers resistant to cucumber mosaic virus, Hort. Environ. Biotechnol. 52(2):211-217, 2011
특허
- •향상된 FISH 수행을 위한 신규한 5S rDNA용 핵산 프로브 및 이를 이용한 신규한 FISH법(10-2017-0096526)
- •향상된 FISH 수행을 위한 신규한 45S rDNA용 핵산 프로브 및 이를 이용한 신규한 FISH법(10-2017-0096527)
저서
- •Quantity, Distribution, and Evolution of Major Repeats in Brassica napus (chapterbook), In: Shengyi Liu, Rod Snowdon, Boulos Chalhoub, The Brassica napus Genome, Compendium of Plant Genomes, 2018, Springer-Verlag, Berlin Heidelberg
- •생명의 위대한 비밀 –유전암호를 풀어라-, 2017, 한국유전학회 공역, ㈜라이프사이언스
- •하트웰의 유전학 –유전자에서 유전체까지-(제5판), 2016, 한국유전학회공역, 홍릉과학출판사
- •Genomic survey of the hidden 7 components of the B. rapa genome (chapterbook), In: X. Wang and C. Kole (eds.), The Brassica rapa Genome, Compendium of Plant Genomes, 2015, Springer-Verlag, Berlin Heidelberg
- •필수유전학(6판, eds.)-유전체전망대에서 바라봄, 2015, 16인 공역, ㈜월드사이언스
- •왓슨 분자생물학(7판, eds.), 2014, 15인 공역, ㈜바이오사이언스출판
- •Johnson’s 생명과학(7판), 2013. 32인 공역, 드림플러스
- •생명과학의 이해, 2013, 29인 공역, 녹문당
- •생명과학, 2012, 38인 공역, 녹문당
- •일반식물학(2판), 2008, 12인 공역, ㈜월드사이언스
- •기초생화학, 2006, 29인 공역, 도서출판 자유아카데미
- •생명과학사전, 2003, 공저, 도서출판 아카데미
-
연구소소개
주요 연구분야
-
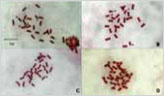
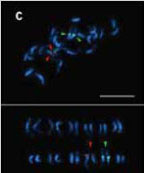
1FISH를 이용한 고추의 핵형분석과 삽입 유전자 CMVPO-CP의 물리적 지도 작성
-
-
-
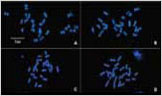
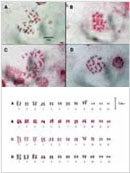
2FISH에 의한 GM 벼의 핵형분석과 transgene의 물리적 지도 작성
-
-
-
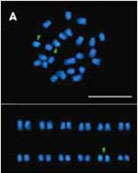
3박과 식물에서 bi-color FISH를 이용한 5S와 45S rDNA의 물리적 유전자 지도 작성
-
-
-
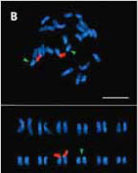
4유전자 변형 작물에서 stacked gene을 검출하기위한 새로운 기술의 개발
-
-
-
5선인장의 돌발바이러스 분자진단용 multiplex real-time PCR 키트 개발 및 무병주 조기선발 시스템 구축
-
-
주요 프로젝트
- • 인삼 및 근연 식물 비교세포유전학 기술 정착 및 염색체분리 기술개발, 농촌진흥청/농생물게놈활용연구사업단과제, 2018. 01. 01~2020. 12. 31
- • FISH 기법에 의한 결명자 유전체 내 반복서열 및 단일서열의 물리지도 작성, 연구재단/기초연구지원사업(중견연구)과제, 2017. 03. 01~2021. 02. 28
- • 배추과 속간 교잡 신품종 개발 및 육종 체계 확립, 농림수산식품기술기획평가원/농생명산업기술개발사업과제, 2017. 04. 01~2019. 12. 31
- • FISH 기법에 의한 결명자 주요 반복서열의 염색체 내 분포, ㈜파이젠, 민간연구용역과제, 2016. 01. 01~ 2016. 12. 31
- • 인삼 유전체의 물리적 지도작성 및 유전체 서열 조립 검정, 농촌진흥청/차세대바이오그린사업, 2015. 01. 15~2017. 12. 31
- • 원예작물 핵형분석 및 유전체 조립지원연구, 농촌진흥청/다부처유전체사업, 2014. 04. 01~2015. 12. 31
- • 농생명자원 유전체 해독 품목별 핵형분석, 농촌진흥청/공동연구사업, 2013.07.01 ~2013. 12. 31
- • FISH 핵형분석 및 5S rDNA 서열에 의한 박과 식물의 계통 유연관계 분석, 연구재단/기초연구사업, 2011. 09. 01~ 2016. 08. 31
- • 인삼 유전체의 분자세포유전학적 분석, 농촌진흥청/차세대바이오그린사업, 2011. 05. 01~2014. 12. 31
- • 선인장의 돌발바이러스 분자진단용 multiplex real-time PCR 키트 개발 및 무병주 조기선발 시스템 구축, 서울지방중소기업청/산학연기술개발사업, 2010. 06. 01~2011. 05. 31
- • Stack gene 검정신기술을 활용한 도입 전자 동태분석용 모니터링 기술 개발, 농촌진흥청/바이오그린21사업, 2008. 03. 01~2010. 12. 31
- • 바이러스내병성 GM 고추의 농업환경위해성 평가, 서울지방중소기업청/산학연기술개발사업, 2007. 04. 01~ 2010. 12. 31
- • 장미신품종육성 및 육종기술 개발, 농진청 원예연구소 장미사업단 사업, 2007. 01. 01~2010. 12. 31.
인력
- 연구원 6명 (박사 1명, 석사 2명, 학사 3명)
보유장비
- •Hybridization Incubator (Combi SV12)
- •Slide Warmer (C-SL)
- •Adjustable Tilt Rocker
- •Waterbath (Lab. Companion 13W-10G)
- •Fluoresence Microscope (Olympus BX51)
- •Refrigerated Waterbath (Eyela UA-1000)
- •UV Transilluminator with Printgraph (ATTA AE-6932GXCF)
- •Centrifugal evaporator (Eyela CVE-2000)
- •Clean bench
- •PCR Thermal Cylcer (Bioer XP Cycler TCP-XP-D)
- •Refrigerated micro highspeed centrifuge (Micro 17TR)
- •Freezer (Samsung)
- •Dry oven
- •General-use incubator
-